Biography
I am a PhD candidate in biomedical engineering at the Johns Hopkins University Institute for Computational Medicine. I am currently working with Dr. Alison Hill and Dr. Feilim Mac Gabhann to build mathematical models and statistical inference frameworks for within-host viral dynamics, integrated with quantitative systems pharmacology models of novel immunotherapies. I received a Bachelor of Science in applied mathematics from the University of Connecticut, where I worked with Dr. Paola Vera-Licona at the Center for Quantitative Medicine to identify putative tumor reversion targets using dynamical systems methods. Following graduation, I spent two years as a post-baccalaureate researcher developing Bayesian methods for systems genetics applications with Dr. Gary Churchill at The Jackson Laboratory in Bar Harbor, Maine. I am fascinated by the use of mathematical and statistical modeling to understand biology, and I foresee a career developing models to aid in the development of novel therapeutic modalities.
Download my CV.
- Systems Genetics
- Personalized Medicine
- Mechanistic Mathematical Modeling
- Systems Pharmacology
- Bayesian Inference
BS in Applied Mathematics, 2020
The University of Connecticut
PhD in Biomedical Engineering, in progress
Johns Hopkins School of Medicine
Skills
Proficient
Proficient
Intermediate
Beginner
Proficient
Intermediate
Intermediate
Intermediate
Proficient
Research Experience
Projects
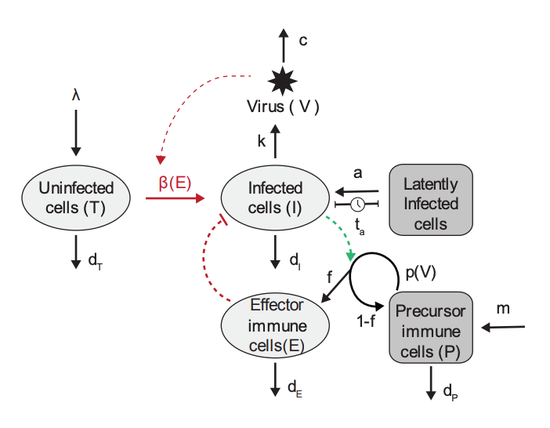
Viral rebound kinetics following single and combination immunotherapy for HIV/SIV
In collaboration with Mélanie Prague at the University of Bordeaux, we developed a mathematical model of within-host HIV dynamics and used nonlinear mixed-effects modeling implemented in Monolix to investigate the mechanism of action of several immunotherapies.
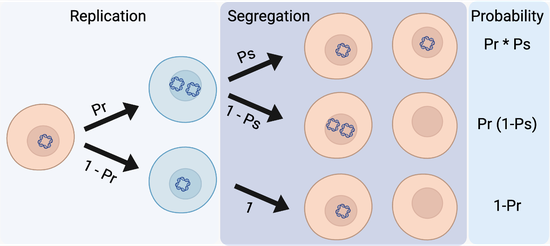
Quantifying the dynamics of Kaposi’s sarcoma-associated herpesvirus persistence
In collaboration with the Kaye lab at Brigham and Women’s Hospital, we are quantifying the dynamics of latent Kaposi’s sarcoma-associated herpesvirus (KSHV) persistence. We developed a mathematical model and a statistical inference framework to infer viral dynamics from fluorescence microscopy images of cells in culture. Forward simulations were used to understand decades-long viral persistence and evaluate latent KSHV replication as a potential therapeutic target to disrupt KSHV-dependent tumor growth.
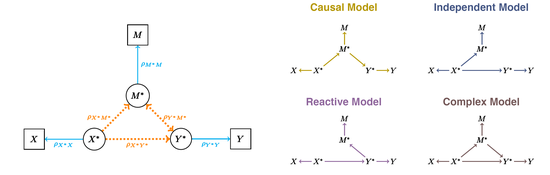
The impact of measurement noise in mediation analysis
As a post-bacc at the Jackson Laboratory, my main research focus has been mediation analysis. A primary objective of mediation analysis is to infer the relationship among three variables, and it is becoming increasingly common to use it with multi-omics data to understand causal pathways underlying a phenotype. Mediation analysis is often done without distinguishing variation due to causal relationships from variation due to measurement noise, which can have a profound effect on inferences. In this analysis, we address the impact of applying a standard mediation analysis to data as if it is measured without error and identify ways to diagnose the accuracy of results from real data.
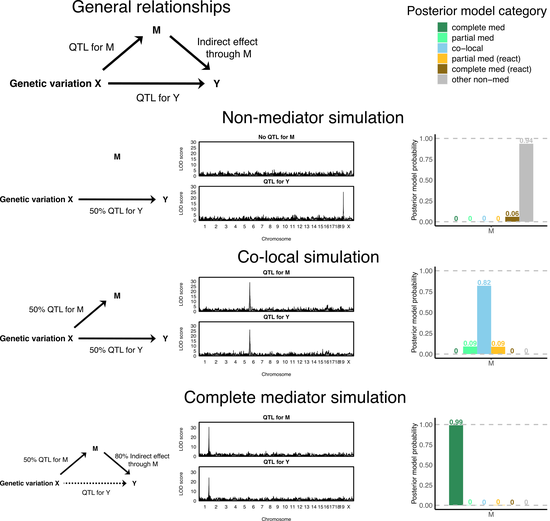
A Bayesian model selection approach to mediation analysis
In collaboration with the Valdar lab at UNC, we developed a Bayesian model selection approach to mediation analysis implemented in the bmediatR R package. This approach allows for flexibility in both data inputs and potential inferences and uses conjugate priors to increase efficiency. I am currently extending the framework to allow for the inference of moderated mediation.
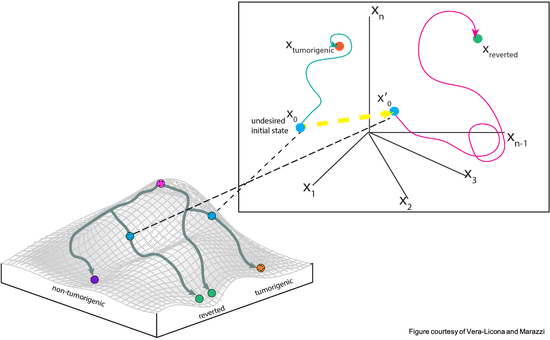
A quantitative pipeline for the identification of combinations of targets for claudin-low triple negative breast cancer reversion
My undergraduate thesis focused on a dynamical approach to identify reversion targets for a subtype of Triple Negative Breast Cancer (TNBC). Through the use of bioinformatics tools, optimal control theory, and machine learning algorithms, several combinations of reversion targets were identified for future validation. My contribution to the project was funded by a Summer Undergraduate Research Fund from the University of Connecticut Office of Undergraduate Research.
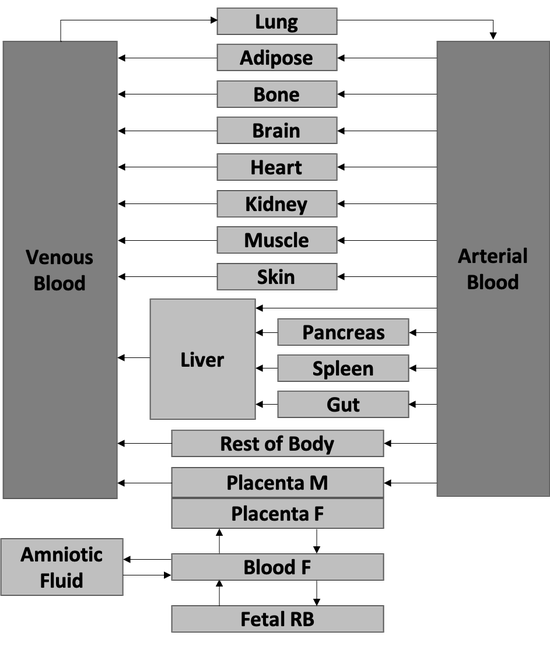
The development of an open and general maternal-fetal physiologically based pharmacokinetic model for drugs metabolized by cytochromes P450 isoenzymes
During a summer internship at Metrum Research Group, I worked with a team of interns to develop a physiologically based pharmacokinetic (PBPK) model for maternal and fetal drug disposition throughout pregnancy. By integrating published data from studies in non-pregnant women with prior knowledge of anatomical, physiological, and biochemical changes associated with pregnancy, we were able to predict drug exposures in a population with limited experimental data. The resulting model can guide dosing prior to drug administration and refine dosing once initial exposures are determined.